rawrr
Web: Bioconductor (https://bioc.r-universe.dev/rawrr)
Installation
We can start with BiocManager::install("rawrr") or install.packages("rawrr", repos = c("https://bioc.r-universe.dev", "https://cloud.r-project.org")) from R, and get the following messages,
> library(rawrr)
'ThermoFisher.CommonCore.BackgroundSubtraction.dll' is missing.
'ThermoFisher.CommonCore.Data.dll' is missing.
'ThermoFisher.CommonCore.MassPrecisionEstimator.dll' is missing.
'ThermoFisher.CommonCore.RawFileReader.dll' is missing.
'ThermoFisher.CommonCore.*.dll' files are not available on the system.
Run 'rawrr::installRawFileReaderDLLs()' or setenv MONO_PATH to the location where the assemblies are located.
For more information, type '?ThermoFisher'.
Package 'rawrr' version 1.12.0 using
RawFileReader reading tool. Copyright © 2016 by Thermo Fisher Scientific, Inc. All rights reserved.
The error messages can be suppressed with installRawFileReaderDLLs()
> if (isFALSE(rawrr::.checkDllInMonoPath())){
+ rawrr::installRawFileReaderDLLs()
+ }
'ThermoFisher.CommonCore.BackgroundSubtraction.dll' is missing.
'ThermoFisher.CommonCore.Data.dll' is missing.
'ThermoFisher.CommonCore.MassPrecisionEstimator.dll' is missing.
'ThermoFisher.CommonCore.RawFileReader.dll' is missing.
'ThermoFisher.CommonCore.*.dll' files are not available on the system.
Run 'rawrr::installRawFileReaderDLLs()' or setenv MONO_PATH to the location where the assemblies are located.
For more information, type '?ThermoFisher'.
Do you accept the Thermo License agreement '/rds/project/rds-4o5vpvAowP0/software/R/rawrr/rawrrassembly/RawFileReaderLicense.txt'? [Y/n]: y
trying URL 'https://github.com/thermofisherlsms/RawFileReader/raw/main/Libs/Net471//ThermoFisher.CommonCore.BackgroundSubtraction.dll'
Content type 'application/octet-stream' length 44544 bytes (43 KB)
==================================================
downloaded 43 KB
trying URL 'https://github.com/thermofisherlsms/RawFileReader/raw/main/Libs/Net471//ThermoFisher.CommonCore.Data.dll'
Content type 'application/octet-stream' length 406016 bytes (396 KB)
==================================================
downloaded 396 KB
trying URL 'https://github.com/thermofisherlsms/RawFileReader/raw/main/Libs/Net471//ThermoFisher.CommonCore.MassPrecisionEstimator.dll'
Content type 'application/octet-stream' length 11264 bytes (11 KB)
==================================================
downloaded 11 KB
trying URL 'https://github.com/thermofisherlsms/RawFileReader/raw/main/Libs/Net471//ThermoFisher.CommonCore.RawFileReader.dll'
Content type 'application/octet-stream' length 654336 bytes (639 KB)
==================================================
downloaded 639 KB
ThermoFisher.CommonCore.BackgroundSubtraction.dll
0
ThermoFisher.CommonCore.Data.dll
0
ThermoFisher.CommonCore.MassPrecisionEstimator.dll
0
ThermoFisher.CommonCore.RawFileReader.dll
0
>
> if (isFALSE(file.exists(rawrr:::.rawrrAssembly()))){
+ rawrr::installRawrrExe()
+ }
>
> library(rawrr)
Package 'rawrr' version 1.12.0 using
RawFileReader reading tool. Copyright © 2016 by Thermo Fisher Scientific, Inc. All rights reserved.
> q()
Testing
We use a real data here,
> cwd <- getwd()
> setwd("~/Caprion/pre_qc_data/spectral_library_ZWK/")
> readSpectrum("szwk901104i19901xms1.raw",scan=1)
[[1]]
Total Ion Current: 10222295
Scan Low Mass: 400
Scan High Mass: 1800
Scan Start Time (min): 0.002808232
Scan Number: 1
Base Peak Intensity: 994858
Base Peak Mass: 445.1201
Scan Mode: FTMS + p NSI Full ms [400.0000-1800.0000]
======= Instrument data ===== : NULL
Multiple Injection: ii
Multi Inject Info: IT=15;15
AGC: On
Micro Scan Count: 1
Scan Segment: 1
Scan Event: 1
Master Index: 0
Charge State: 0
Monoisotopic M/Z: 0.0000
Ion Injection Time (ms): 30.000
Max. Ion Time (ms): 30.00
FT Resolution: 70000
MS2 Isolation Width: 1400.00
MS2 Isolation Offset: 0.00
AGC Target: 3000000
HCD Energy:
Analyzer Temperature: 29.92
=== Mass Calibration: NULL
Conversion Parameter B: 67839603.0252
Conversion Parameter C: 35794268.3879
Temperature Comp. (ppm): -5.61
RF Comp. (ppm): 0.08
Space Charge Comp. (ppm): -0.22
Resolution Comp. (ppm): -0.15
Number of Lock Masses: 0
Lock Mass #1 (m/z): 0.0000
Lock Mass #2 (m/z): 0.0000
Lock Mass #3 (m/z): 0.0000
LM Search Window (ppm): 0.0
LM Search Window (mmu): 0.0
Number of LM Found: 0
Last Locking (sec): 0.0
LM m/z-Correction (ppm): 0.00
=== Ion Optics Settings: NULL
S-Lens RF Level: 50.00
S-Lens Voltage (V): 25.00
Skimmer Voltage (V): 15.00
Inject Flatapole Offset (V): 8.00
Bent Flatapole DC (V): 6.00
MP2 and MP3 RF (V): 900.00
Gate Lens Voltage (V): 5.25
C-Trap RF (V): 2400.0
==== Diagnostic Data: NULL
Intens Comp Factor: 0.6884
Res. Dep. Intens: 1.215
CTCD NumF: 16
CTCD Comp: 5.390
CTCD ScScr: 0.792
RawOvFtT: 231010.0
LC FWHM parameter: 15.0
Rod: 0
PS Inj. Time (ms): 0.640
AGC PS Mode: 1
AGC PS Diag: 4001800
HCD Energy eV: 0.000
AGC Fill: 0.01
Injection t0: 0.044
t0 FLP: 429.09
Access Id: 0
Analog Input 1 (V): 0.000
Analog Input 2 (V): 0.000
attr(,"class")
[1] "rawrrSpectrumSet"
> sz1 <- readSpectrum("szwk901104i19901xms1.raw",scan=1)
x$centroidStream is not TRUE
> setwd(cwd)
> png("sz.png",res=300,height=6,width=6,units="in")
> plot(sz1[[1]], centroid=FALSE)
> dev.off()
as shown here, 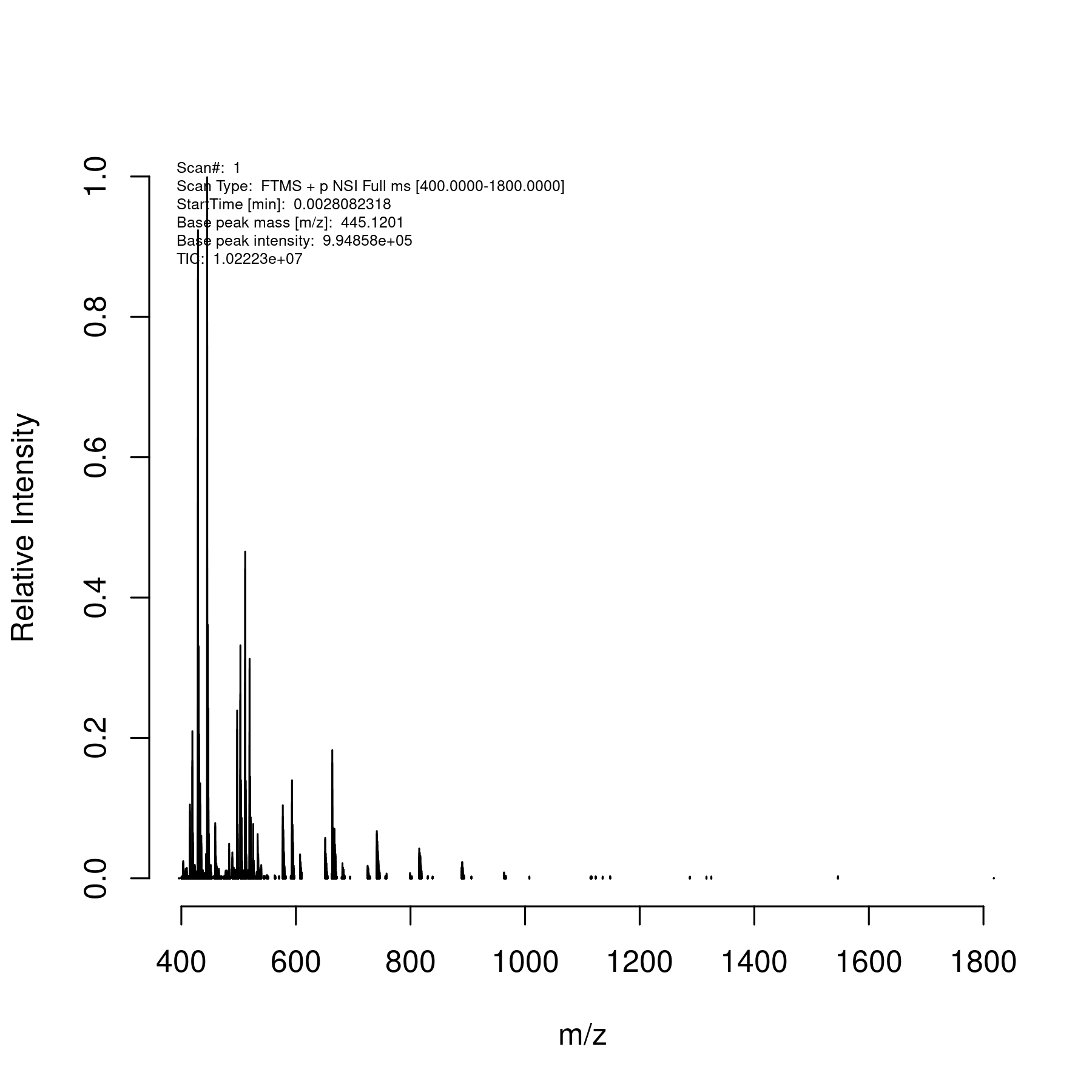 .
.
MsBackendRawFileReader
This is closely related; for the example above we try
> beRaw <- Spectra::backendInitialize(MsBackendRawFileReader::MsBackendRawFileReader(),
+ files = dir(patt="raw"))
> class(beRaw)
[1] "MsBackendRawFileReader"
attr(,"package")
[1] "MsBackendRawFileReader"
> beRaw
MsBackendRawFileReader with 1935241 spectra
msLevel rtime scanIndex
<integer> <numeric> <integer>
1 1 0.00280752 1
2 2 0.00977762 2
3 2 0.01136105 3
4 1 0.01378255 4
5 1 0.01998046 5
... ... ... ...
1935237 1 52.4772 24262
1935238 1 52.4829 24263
1935239 1 52.4886 24264
1935240 1 52.4942 24265
1935241 1 52.4999 24266
... 30 more variables/columns.
file(s):
szwk021704i19101xms1.raw
szwk021704i19101xms2.raw
szwk021704i19101xms3.raw
... 77 more files
> beRaw |> Spectra::spectraVariables()
[1] "msLevel" "rtime"
[3] "acquisitionNum" "scanIndex"
[5] "mz" "intensity"
[7] "dataStorage" "dataOrigin"
[9] "centroided" "smoothed"
[11] "polarity" "precScanNum"
[13] "precursorMz" "precursorIntensity"
[15] "precursorCharge" "collisionEnergy"
[17] "isolationWindowLowerMz" "isolationWindowTargetMz"
[19] "isolationWindowUpperMz" "scanType"
[21] "charge" "masterScan"
[23] "dependencyType" "monoisotopicMz"
[25] "AGC" "injectionTime"
[27] "resolution" "isolationWidth"
[29] "isolationOffset" "AGCTarget"
[31] "collisionEnergyList" "AGCFill"
[33] "isStepped"
See MsBackendRawFileReader.html for additional information.
