fsc2
Web: https://cmpg.unibe.ch/software/fastsimcoal2/
Installation
wget https://cmpg.unibe.ch/software/fastsimcoal2/downloads/fsc28_linux64.zip
unzip fsc28_linux64.zip
cd fsc28_linux64/
chmod +x fsc28
./fsc28
which gives command line options.
fsc was launched without any argument: attempting to read file "fsc_run.txt"
Unable to find file "fsc_run.txt" in current directory(/usr/local/Cluster-Apps/ceuadmin/fsc2/2.8.0/example files)
fastSimcoal2 (ver 2.8.0.0 - 22.09.23)
Usage:
-h --help : prints this help
-i --ifile test.par : name of parameter file
-n --numsims 1000 : number of simulations to perform
Also applies for parameter estimation
-t --tplfile test.tpl : name of template parameter file (optional)
-f --dfile test.def : name of parameter definition file (optional)
-F --dFile test.def : same as -f, but only uses simple parameters defined
in template file. Complex params are recomputed
-e --estfile test.est : parameter prior definition file (optional)
Parameters drawn from specified distributions are
substituted into template file.
-E --numest 10 : number of draws from parameter priors (optional)
Listed parameter values are substituted in template file
-g --genotypic : generates arlequin projects with genotypic data
-p --phased : specifies that phase is known in arlequin output
default: phase is unknown
-s --dnatosnp 2000 : output DNA as SNP data, and specify maximum no. of SNPs
to output (use 0 to output all SNPs).
-S --allsites : output the whole DNA sequence, incl. monomorphic sites
-I --inf : generates DNA mutations according to an
infinite site (IS) mutation model
-d --dsfs : computes derived site frequency spectrum
(for SNP or DNA as SNP (-s) data only).
-m --msfs : computes minor site frequency spectrum
(for SNP or DNA as SNP (-s) data only)
-j --jobs : output one simulated or bootstrapped SFS per file
in a separate directory for easier analysis
(requires -d or -m and -s0 options)
-b --numboot 10 : number of bootstraps to perform on polymorphic sites to extract SFS
(should be used in addition to -s0 and -j options)
-H --header : generates header in site frequency spectrum files
-q --quiet : minimal message output to console
-T --tree : outputs coalescent tree in nexus format
-k --keep 10000 : number of simulated polymorphic sites kept in memory
If the simulated no. is larger, then temporary files
are created. Default value is 10000
-K --numRandGen 20000 : number of random numbers generated in advance
Default value is 20000
-r --seed : seed for random number generator (positive integer <= 1E6)
-x --noarloutput : does not generate Arlequin output
-G --indgenot : generates an individual genotype table
-M --maxlhood : perform parameter estimation by max lhood from SFS
values between iterations
-L --numloops 20 : number of loops (ECM cycles) to perform during
lhood maximization. Default is 20
-l --minnumloops 2 : number of loops (ECM cycles) for which the lhood is
computed on both monomorphic and polymorphic sites
if REFERENCE parameter is defined
-C --minSFSCount 1 : minimum observed SFS entry count taken into account in
likelihood computation (default = 1, but value can be < 1. e.g 0.5)
-0 --removeZeroSFS : do not take into account monomorphic sites for SFS
likelihood computation
-a --ascDeme 0 : This is the deme id where ascertainment is performed
when simulating SNPs. Default: no ascertainment.
-A --ascSize 2 : number of ascertained chromosomes used to define SNPs in
a given deme. Optional parameter. Default value is 2
-u --multiSFS : generate or use multidimensional SFS
-w --brentol 0.01 : tolerance for Brent optimization
Default = 0.01. Smaller value imply more precise estimations
but require more computation time (min;max) = (1e-1;1e-5)
-c --cores 1 : number of openMP threads for parameter estimation
(default=1, max=numBatches, use 0 to let openMP choose optimal value)
-B --numBatches 12 : max. no. of batches for multi-threaded runs
(default=12)
-P --pooledsfs : computes pooled SFS over all samples.
Assumes -d or -m, but not -u flag activated
--recordMRCA : records tMRCAs for each non recombining segment and outputs
results in <generic name>_mrca.txt. Beware: huge slow down of computing time
--foldedSFS : computes the 1D and 2D MAF SFS by simply folding the DAF SFS
--logprecision 23 : precision for computation of logs of random numbers. Max value is 23
Default value is 23 (full precision). Recommended lower value is 18
--initValues my.pv : specifies a file (*.pv) containing initial parameter values
for parameter optimization
--nosingleton : ignores singletons in likelihood computation
-y --resetParam 3 : Number of unsuccessful cycles before resetting parameters to current max lhood values
default is zero, implying no resetting
-z --finalRange 0.01 : Proportion of the initial search range remaining in the last cycle (default is 1)
Testing
When the module is built, one can use this script.
module load ceuadmin/fsc2/2.8.0
cd examples
fsc28 -i 1PopDNA.par -n 1 -d -e
module load ceadmin/R
wget https://cmpg.unibe.ch/software/fastsimcoal2/R/ParFileViewer.r
Rscript ParFileViewer.r 1PopDNA.par
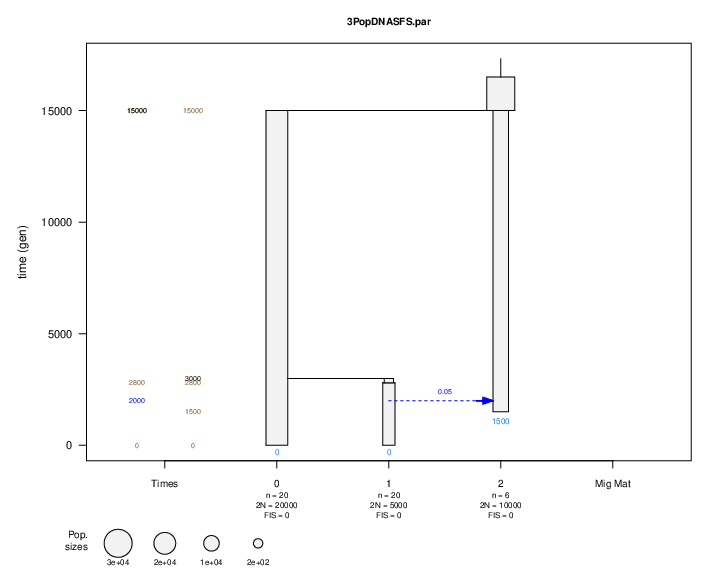
Rscript ParFileViewer.r 3PopDNASFS.par
convert 3PopDNASFS.par.pdf 3PopDNASFS.png
where an R utility is used to visually inspect the validity of modeled scenarios, 3PopDNASFS.png.
References
Excoffier L, et al. fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics 37 (24):4882–4885, 2021.
