Relate
Web: https://myersgroup.github.io/relate/
1.2.3
Firefox / 142.0 would lose part of the files, and Chrome is used instead. Both dynamic and static distributions work well under CSD3.
Besides defining an evironmental variable PATH_TO_RELATE, ceuadmin/R/latest is also called in the module definition.
Testing
A well-organised run-relate.sh can be started from example/ directory; the last two code chunks are used here.
module load ceuadmin/relate
cd example
${PATH_TO_RELATE}/scripts/SampleBranchLengths/SampleBranchLengths.sh \
-i example \
-o example_bypop_sampled \
-m 1.25e-8 \
--coal example_bypop.coal \
--first_bp 199990000 \
--last_bp 200010000 \
--seed 1 \
--num_samples 100
${PATH_TO_RELATE}/scripts/TreeView/TreeViewSample.sh \
--haps ./data/example.haps.gz \
--sample ./data/example.sample.gz \
--anc example_bypop_sampled.anc \
--mut example_bypop_sampled.mut \
--bp_of_interest 200000000 \
--years_per_gen 28 \
--poplabels ./data/example.poplabels \
--dist example_bypop_sampled.dist \
-o plot
convert plot.pdf relate-plot.png
where the module mirrors the definition in run-relate.sh with the named global variable PATH_TO_RELATE. Internally, the second chunk here calls R/ggplot2 for plot.pdf, which is converted by ImageMagick to relate-plot.png
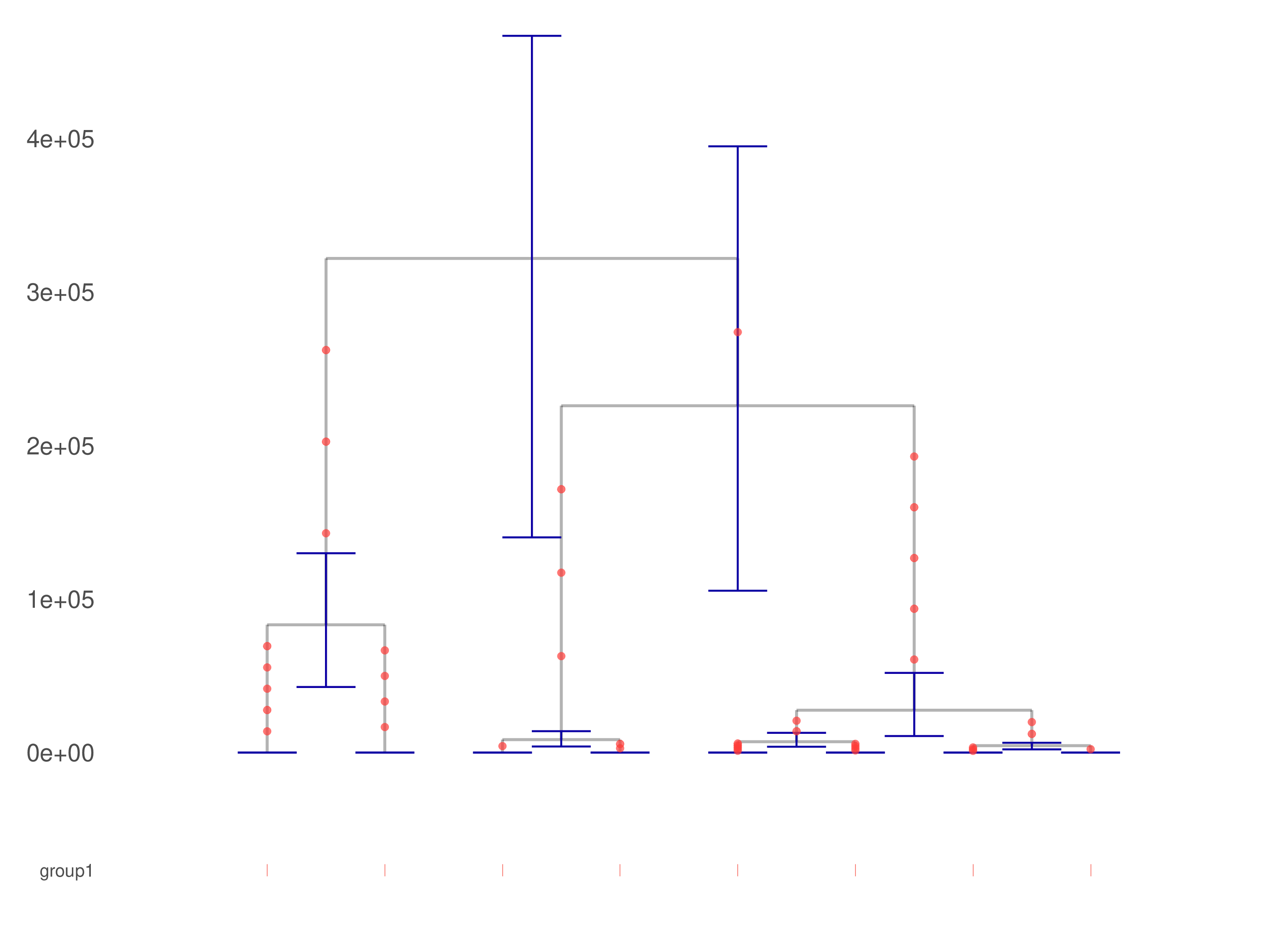
and shown above.
References
Liu X, et al. Selection at the GSDMC locus in horses and its implications for human mobility. Science 389: 925-930 (2025). https://www.science.org/doi/10.1126/science.adp4581.
Speidel, L, et al. A method for genome-wide genealogy estimation for thousands of samples. Nat Genet 51: 1321–1329 (2019). https://doi.org/10.1038/s41588-019-0484-x
Speidel L, et al., Inferring population histories for ancient genomes Using genome-Wide genealogies. Mol Biol Evol 38 (9): 3497–3511, 2021, https://doi.org/10.1093/molbev/msab174
